Introduction
Psychometric Network Analysis (PNA), which is also referred to as “network psychometrics”, is an emerging field that combines concepts from psychometrics and network analysis to study the relationships among psychological variables. Generally, psychometrics involves the measurement of latent traits, while network analysis focuses on modeling and analyzing complex systems of interconnected elements. PNA aims to represent the relationships between psychological constructs, such as symptoms, traits, or behaviors, as a network of interconnected nodes. The figure below shows a network graph for the Big Five Personality Test:
](pna1.png)
Figure 1: Figure from Epskamp (2017)
Each node represents a specific personality trait, and the edges (connections) between nodes reflect the statistical relationships between those variables. The edges can be weighted to represent the strength (e.g., thicker edges represents stronger associations) and direction of the relationships between the corresponding traits (e.g., green lines for positive and red lines for negative associations).
In social sciences (especially in psychology and education), researchers often use “partial correlation” coefficients to understand the associations between the variables. A partial correlation is the association between two [quantitative] variables, after conditioning on all other variables in the dataset. Networks based on partial correlations are known as Gaussian Graphical Models (GGMs; Costantini et al. (2015)). A GGM can be mathematically expressed as follows:
\[Σ=Δ(I−Ω)^{−1}Δ,\] where \(Σ\) is the variance-covariance matrix of the observed variables, \(Δ\) is a diagonal matrix, \(Ω\) is a matrix containing weights linking observed variables, conditioned on all other variables, and \(I\) is the identity matrix.
To prevent over-interpretation in network structures, we want to limit the number of spurious connections (i.e., weak partial correlations due to sampling variation). It is possible to eliminate spurious connections using statistical regularization techniques, such as “least absolute shrinkage and selection operator” (LASSO; Tibshirani (1996)). LASSO (L1) regularization shrinks partial correlation coefficients when estimating a network model, which means that small coefficients are estimated to be exactly zero.
As an extension of LASSO, graphical LASSO, or shortly glasso (Friedman et al., 2007), involves a penalty parameter (\(\lambda\)) to remove weak associations within a network. The following figure demonstrates the impact of the penalty parameter.
](pna2.png)
Figure 2: Figure Adapted from Epskamp and Fried (2017)
When estimating network models, glasso is typically combined with the extended Bayesian information criterion (EBIC; Chen & Chen (2008)) for tuning parameter selection, resulting in EBICglasso (Epskamp & Fried, 2018). If the dataset for PNA consists of continuous variables that are multivariate normally distributed, then we can estimate a GGM based on partial correlations with glasso or EBICglasso. GGM can also be used for ordinal data (e.g., Likert-scale data) wherein the network is based on the polychoric correlations instead of partial correlations.
In this blog post, I want to demonstrate how to perform PNA and visualize resulting network models (specifically, GGMs) using R 🕸 👩💻. I highly encourage readers to check out Network Psychometrics with R by Isvoranu et al. (2022) for a comprehensive discussion of network models and their estimation using R.
Let’s get started 💪.
Example
In our example, we will use a subset of the Synthetic Aperture Personality Assessment (SAPA)–a web based personality assessment project (https://www.sapa-project.org/). The purpose of SAPA is to find patterns among the vast number of ways that people differ from one another in terms of their thoughts, feelings, interests, abilities, desires, values, and preferences (Condon & Revelle, 2014; Revelle et al., 2010). The dataset consists of 16 SAPA items sampled from the full instrument (80 items). These items measure four subskills (i.e., verbal reasoning, letter series, matrix reasoning, and spatial rotations) as part of the general intelligence, also known as g factor. The “sapa_data.csv” dataset is a data frame with 1525 individuals who responded to 16 multiple-choice items in SAPA. The original dataset is included in the psych package (William Revelle, 2023). The dataset can be downloaded from here. Now, let’s import the data into R and then review its content.
Next, we will check out the correlations among the SAPA items. Since the items are dichotomous (i.e., binary), we will compute the tetrachoric correlations and then visualize the matrix using a correlation matrix plot.
library("psych")
library("ggcorrplot")
# Save the correlation matrix
cormat <- psych::tetrachoric(x = sapa)$rho# Correlation matrix plot
ggcorrplot::ggcorrplot(corr = cormat, # correlation matrix
type = "lower", # print only the lower part of the correlation matrix
hc.order = TRUE, # hierarchical clustering
show.diag = TRUE, # show the diagonal values of 1
lab = TRUE, # add correlation values as labels
lab_size = 3) # Size of the labels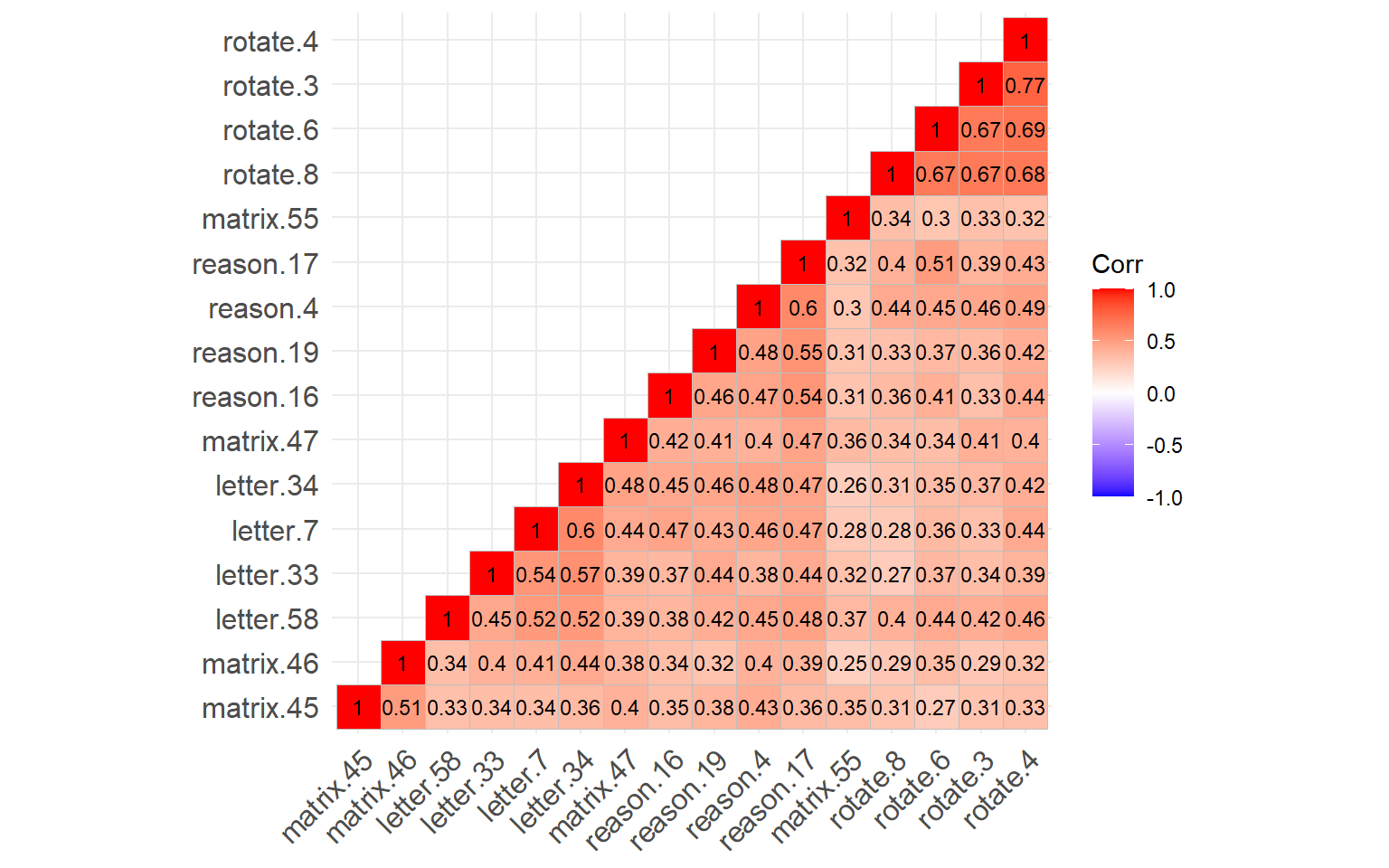
Figure 3: Correlation matrix plot of the SAPA items
The figure above shows that all of the SAPA items are positively correlated with each other. the items associated with the same construct (e.g., rotation) seem to have been clustered together (e.g., see the rotate.4, rotate.3, rotate.6, and rotate.8 as the top cluster). Now, we can go ahead and estimate a GGM based on this correlation matrix.
We will use the bootnet package (Epskamp, Borsboom, et al., 2018). This package has many methods to estimate GGMs. In our example, we will use “IsingFit” because the Ising model deal with binary data (see Epskamp, Maris, et al. (2018) and Marsman et al. (2018) for more details on the Ising network models). This model combines L1-regularized logistic regression with model selection based on EBIC.
library("bootnet")
network_sapa_1 <- bootnet::estimateNetwork(
data = sapa, # dataset
default = "IsingFit", # for estimating GGM with the Ising model and EBIC,
verbose = FALSE # Ignore real-time updates on the estimation process
)
# View the estimated network
plot(network_sapa_1, layout = "spring") 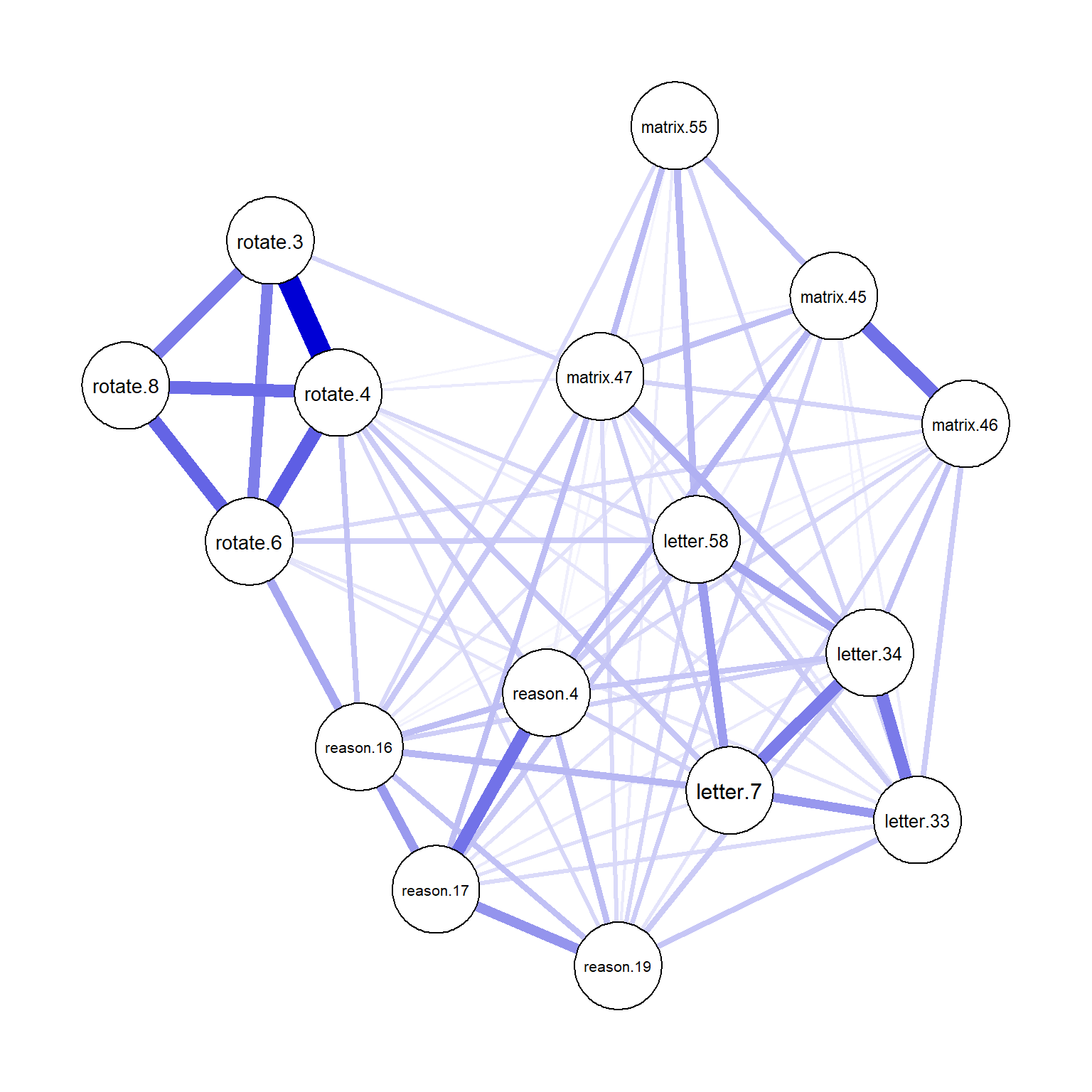
Figure 4: Network plot for the Ising model
From the network plot above, we can see that the items associated with the same construct often have stronger edges between the nodes (e.g., see the thick edges between the rotation items). These items also appear to be clustered closely within the network.
What if we compute the polychoric correlations for the SAPA items using corMethod = "cor_auto" and estimate a GGM with glasso and EBIC?
network_sapa_2 <- bootnet::estimateNetwork(
data = sapa,
corMethod = "cor_auto", # for polychoric correlations
default = "EBICglasso", # for estimating GGM with glasso and EBIC
verbose = FALSE
)
# View the estimated network
plot(network_sapa_2, layout = "spring") 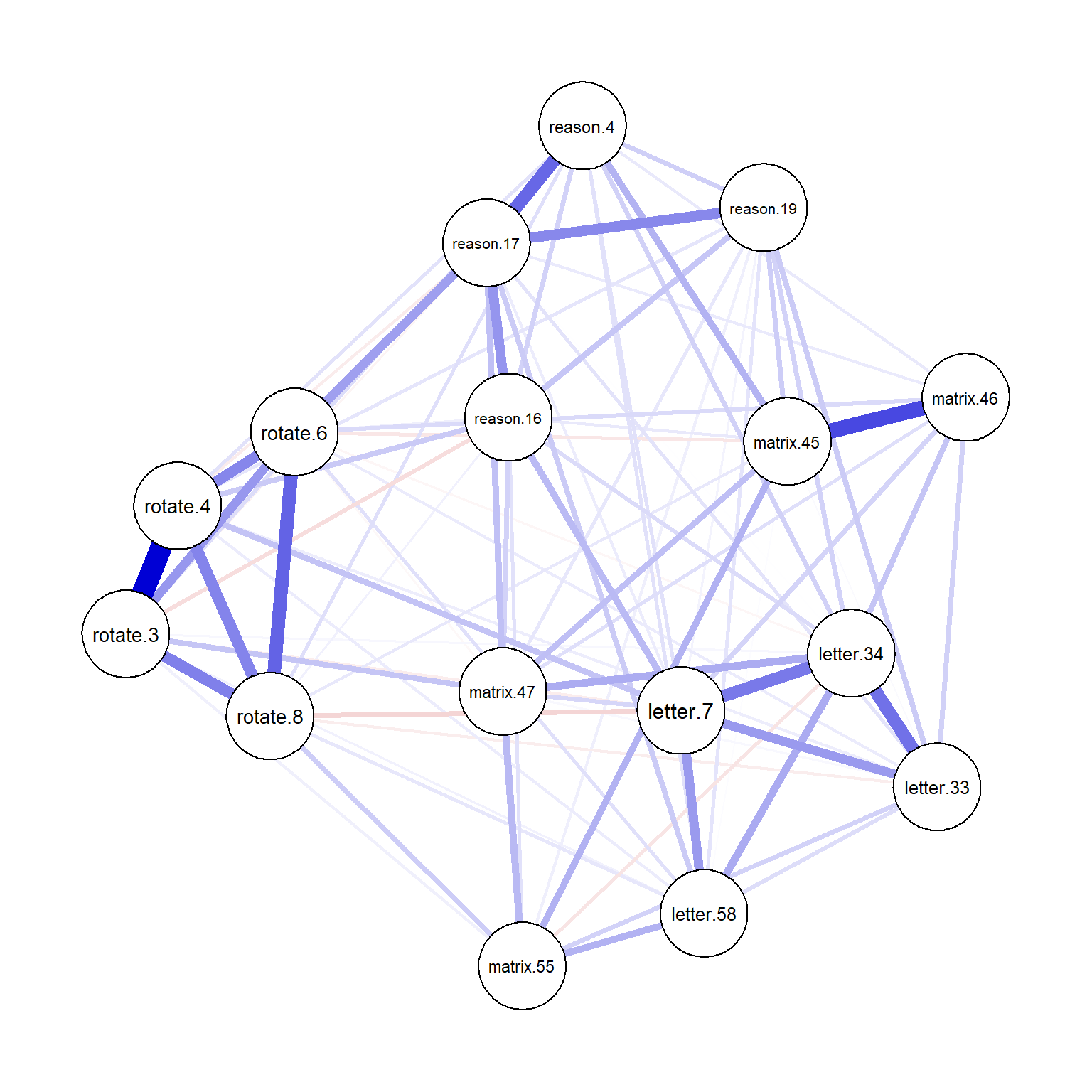
Figure 5: Network plot for the GGM
By default, the bootnet package uses tuning = 0.5 for the penalty parameter for EBICglasso. We can increase or decrease this parameter to adjust the penalty on the model (Note: tuning = 0 leads to model selection based on BIC instead of EBIC). In the following example, we will use tuning = 1 to apply a larger penalty parameter to the model.
network_sapa_3 <- bootnet::estimateNetwork(
data = sapa,
corMethod = "cor_auto", # for polychoric correlations
default = "EBICglasso", # for estimating GGM with glasso and EBIC
tuning = 1,
verbose = FALSE
)
# View the estimated network
plot(network_sapa_3, layout = "spring") 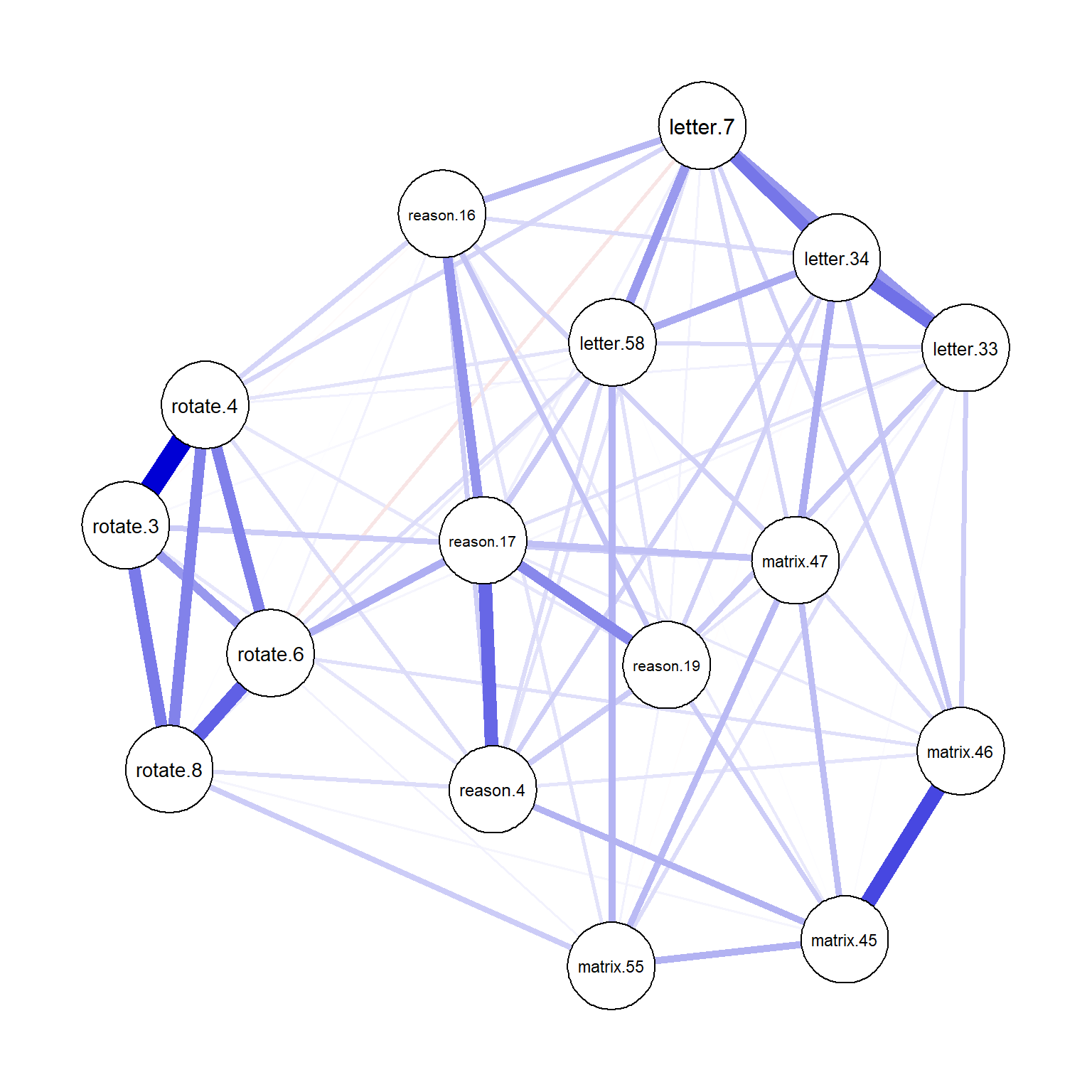
Figure 6: Network plot for the GGM after tuning
The final network plot includes fewer edges compared to the previous one with the default tuning parameter of 0.5. As the tuning parameter gets larger, more edges may be eliminated from the network, leading to a more compact and interpretable model.
I also want to demonstrate the same analysis using the psychonetrics package (Epskamp, 2023). This package is more versatile than bootnet as it is capable of estimating both network models and structural equation models.
library("psychonetrics")
library("dplyr")
network_sapa_4 <- psychonetrics::ggm(data = sapa,
omega = "full", # or "empty" to set all elements to zero
delta = "full", # or "empty" to set all elements to zero
estimator = "FIML", # or "ML", "ULS", or "DWLS"
verbose = FALSE) %>%
psychonetrics::runmodel() # Run the model
# View the model parameters
network_sapa_4 %>%
psychonetrics::parameters()
Parameters for group fullsample
- mu
var1 op var2 est se p row col par
reason.4 ~1 0.64 0.012 < 0.0001 1 1 1
reason.16 ~1 0.70 0.012 < 0.0001 2 1 2
reason.17 ~1 0.70 0.012 < 0.0001 3 1 3
reason.19 ~1 0.62 0.012 < 0.0001 4 1 4
letter.7 ~1 0.60 0.013 < 0.0001 5 1 5
letter.33 ~1 0.57 0.013 < 0.0001 6 1 6
letter.34 ~1 0.61 0.012 < 0.0001 7 1 7
letter.58 ~1 0.44 0.013 < 0.0001 8 1 8
matrix.45 ~1 0.53 0.013 < 0.0001 9 1 9
matrix.46 ~1 0.55 0.013 < 0.0001 10 1 10
matrix.47 ~1 0.61 0.012 < 0.0001 11 1 11
matrix.55 ~1 0.37 0.012 < 0.0001 12 1 12
rotate.3 ~1 0.19 0.010 < 0.0001 13 1 13
rotate.4 ~1 0.21 0.010 < 0.0001 14 1 14
rotate.6 ~1 0.30 0.012 < 0.0001 15 1 15
rotate.8 ~1 0.19 0.0099 < 0.0001 16 1 16
- omega (symmetric)
var1 op var2 est se p row col par
reason.16 -- reason.4 0.086 0.025 0.00072 2 1 17
reason.17 -- reason.4 0.20 0.025 < 0.0001 3 1 18
reason.19 -- reason.4 0.091 0.025 0.00034 4 1 19
letter.7 -- reason.4 0.064 0.026 0.012 5 1 20
letter.33 -- reason.4 0.0015 0.026 0.95 6 1 21
letter.34 -- reason.4 0.077 0.025 0.0024 7 1 22
letter.58 -- reason.4 0.060 0.026 0.018 8 1 23
matrix.45 -- reason.4 0.10 0.025 < 0.0001 9 1 24
matrix.46 -- reason.4 0.054 0.026 0.033 10 1 25
matrix.47 -- reason.4 0.025 0.026 0.33 11 1 26
matrix.55 -- reason.4 0.013 0.026 0.61 12 1 27
rotate.3 -- reason.4 0.037 0.026 0.14 13 1 28
rotate.4 -- reason.4 0.034 0.026 0.18 14 1 29
rotate.6 -- reason.4 0.035 0.026 0.17 15 1 30
rotate.8 -- reason.4 0.034 0.026 0.19 16 1 31
reason.17 -- reason.16 0.15 0.025 < 0.0001 3 2 32
reason.19 -- reason.16 0.090 0.025 0.00041 4 2 33
letter.7 -- reason.16 0.097 0.025 0.00014 5 2 34
letter.33 -- reason.16 0.018 0.026 0.48 6 2 35
letter.34 -- reason.16 0.066 0.026 0.010 7 2 36
letter.58 -- reason.16 0.0081 0.026 0.75 8 2 37
matrix.45 -- reason.16 0.042 0.026 0.098 9 2 38
matrix.46 -- reason.16 0.021 0.026 0.41 10 2 39
matrix.47 -- reason.16 0.073 0.025 0.0039 11 2 40
matrix.55 -- reason.16 0.046 0.026 0.074 12 2 41
rotate.3 -- reason.16 -0.027 0.026 0.30 13 2 42
rotate.4 -- reason.16 0.040 0.026 0.12 14 2 43
rotate.6 -- reason.16 0.043 0.026 0.094 15 2 44
rotate.8 -- reason.16 0.020 0.026 0.43 16 2 45
reason.19 -- reason.17 0.15 0.025 < 0.0001 4 3 46
letter.7 -- reason.17 0.049 0.026 0.055 5 3 47
letter.33 -- reason.17 0.055 0.026 0.031 6 3 48
letter.34 -- reason.17 0.037 0.026 0.14 7 3 49
letter.58 -- reason.17 0.068 0.026 0.0079 8 3 50
matrix.45 -- reason.17 0.0052 0.026 0.84 9 3 51
matrix.46 -- reason.17 0.046 0.026 0.075 10 3 52
matrix.47 -- reason.17 0.092 0.025 0.00030 11 3 53
matrix.55 -- reason.17 0.017 0.026 0.50 12 3 54
rotate.3 -- reason.17 -0.011 0.026 0.66 13 3 55
rotate.4 -- reason.17 -0.021 0.026 0.40 14 3 56
rotate.6 -- reason.17 0.092 0.025 0.00030 15 3 57
rotate.8 -- reason.17 0.012 0.026 0.64 16 3 58
letter.7 -- reason.19 0.045 0.026 0.078 5 4 59
letter.33 -- reason.19 0.081 0.025 0.0015 6 4 60
letter.34 -- reason.19 0.074 0.025 0.0039 7 4 61
letter.58 -- reason.19 0.054 0.026 0.035 8 4 62
matrix.45 -- reason.19 0.076 0.025 0.0030 9 4 63
matrix.46 -- reason.19 0.00040 0.026 0.99 10 4 64
matrix.47 -- reason.19 0.055 0.026 0.031 11 4 65
matrix.55 -- reason.19 0.036 0.026 0.16 12 4 66
rotate.3 -- reason.19 0.013 0.026 0.60 13 4 67
rotate.4 -- reason.19 0.034 0.026 0.19 14 4 68
rotate.6 -- reason.19 0.014 0.026 0.57 15 4 69
rotate.8 -- reason.19 0.0054 0.026 0.83 16 4 70
letter.33 -- letter.7 0.14 0.025 < 0.0001 6 5 71
letter.34 -- letter.7 0.18 0.025 < 0.0001 7 5 72
letter.58 -- letter.7 0.13 0.025 < 0.0001 8 5 73
matrix.45 -- letter.7 0.013 0.026 0.60 9 5 74
matrix.46 -- letter.7 0.070 0.025 0.0063 10 5 75
matrix.47 -- letter.7 0.072 0.025 0.0049 11 5 76
matrix.55 -- letter.7 0.0072 0.026 0.78 12 5 77
rotate.3 -- letter.7 -0.018 0.026 0.48 13 5 78
rotate.4 -- letter.7 0.060 0.026 0.019 14 5 79
rotate.6 -- letter.7 0.013 0.026 0.63 15 5 80
rotate.8 -- letter.7 -0.031 0.026 0.23 16 5 81
letter.34 -- letter.33 0.18 0.025 < 0.0001 7 6 82
letter.58 -- letter.33 0.070 0.025 0.0059 8 6 83
matrix.45 -- letter.33 0.030 0.026 0.25 9 6 84
matrix.46 -- letter.33 0.075 0.025 0.0034 10 6 85
matrix.47 -- letter.33 0.038 0.026 0.14 11 6 86
matrix.55 -- letter.33 0.063 0.026 0.013 12 6 87
rotate.3 -- letter.33 0.013 0.026 0.61 13 6 88
rotate.4 -- letter.33 0.029 0.026 0.26 14 6 89
rotate.6 -- letter.33 0.044 0.026 0.087 15 6 90
rotate.8 -- letter.33 -0.025 0.026 0.32 16 6 91
letter.58 -- letter.34 0.11 0.025 < 0.0001 8 7 92
matrix.45 -- letter.34 0.023 0.026 0.37 9 7 93
matrix.46 -- letter.34 0.084 0.025 0.00092 10 7 94
matrix.47 -- letter.34 0.11 0.025 < 0.0001 11 7 95
matrix.55 -- letter.34 -0.027 0.026 0.30 12 7 96
rotate.3 -- letter.34 0.024 0.026 0.35 13 7 97
rotate.4 -- letter.34 0.014 0.026 0.59 14 7 98
rotate.6 -- letter.34 -0.014 0.026 0.58 15 7 99
rotate.8 -- letter.34 -0.0020 0.026 0.94 16 7 100
matrix.45 -- letter.58 0.026 0.026 0.32 9 8 101
matrix.46 -- letter.58 0.021 0.026 0.42 10 8 102
matrix.47 -- letter.58 0.026 0.026 0.31 11 8 103
matrix.55 -- letter.58 0.10 0.025 < 0.0001 12 8 104
rotate.3 -- letter.58 0.032 0.026 0.21 13 8 105
rotate.4 -- letter.58 0.046 0.026 0.073 14 8 106
rotate.6 -- letter.58 0.069 0.026 0.0065 15 8 107
rotate.8 -- letter.58 0.039 0.026 0.12 16 8 108
matrix.46 -- matrix.45 0.22 0.024 < 0.0001 10 9 109
matrix.47 -- matrix.45 0.089 0.025 0.00049 11 9 110
matrix.55 -- matrix.45 0.10 0.025 < 0.0001 12 9 111
rotate.3 -- matrix.45 0.018 0.026 0.49 13 9 112
rotate.4 -- matrix.45 0.017 0.026 0.51 14 9 113
rotate.6 -- matrix.45 -0.027 0.026 0.29 15 9 114
rotate.8 -- matrix.45 0.032 0.026 0.21 16 9 115
matrix.47 -- matrix.46 0.063 0.026 0.013 11 10 116
matrix.55 -- matrix.46 0.011 0.026 0.68 12 10 117
rotate.3 -- matrix.46 -0.0057 0.026 0.82 13 10 118
rotate.4 -- matrix.46 0.0016 0.026 0.95 14 10 119
rotate.6 -- matrix.46 0.062 0.026 0.015 15 10 120
rotate.8 -- matrix.46 0.013 0.026 0.61 16 10 121
matrix.55 -- matrix.47 0.089 0.025 0.00045 12 11 122
rotate.3 -- matrix.47 0.054 0.026 0.036 13 11 123
rotate.4 -- matrix.47 0.017 0.026 0.50 14 11 124
rotate.6 -- matrix.47 ~ 0 0.026 1.0 15 11 125
rotate.8 -- matrix.47 0.014 0.026 0.58 16 11 126
rotate.3 -- matrix.55 0.038 0.026 0.14 13 12 127
rotate.4 -- matrix.55 0.014 0.026 0.59 14 12 128
rotate.6 -- matrix.55 0.019 0.026 0.45 15 12 129
rotate.8 -- matrix.55 0.063 0.026 0.013 16 12 130
rotate.4 -- rotate.3 0.33 0.023 < 0.0001 14 13 131
rotate.6 -- rotate.3 0.16 0.025 < 0.0001 15 13 132
rotate.8 -- rotate.3 0.19 0.025 < 0.0001 16 13 133
rotate.6 -- rotate.4 0.19 0.025 < 0.0001 15 14 134
rotate.8 -- rotate.4 0.19 0.025 < 0.0001 16 14 135
rotate.8 -- rotate.6 0.20 0.025 < 0.0001 16 15 136
- delta (diagonal)
var1 op var2 est se p row col par
reason.4 ~/~ reason.4 0.41 0.0074 < 0.0001 1 1 137
reason.16 ~/~ reason.16 0.40 0.0073 < 0.0001 2 2 138
reason.17 ~/~ reason.17 0.38 0.0070 < 0.0001 3 3 139
reason.19 ~/~ reason.19 0.42 0.0077 < 0.0001 4 4 140
letter.7 ~/~ letter.7 0.41 0.0075 < 0.0001 5 5 141
letter.33 ~/~ letter.33 0.43 0.0078 < 0.0001 6 6 142
letter.34 ~/~ letter.34 0.40 0.0073 < 0.0001 7 7 143
letter.58 ~/~ letter.58 0.43 0.0078 < 0.0001 8 8 144
matrix.45 ~/~ matrix.45 0.44 0.0081 < 0.0001 9 9 145
matrix.46 ~/~ matrix.46 0.44 0.0080 < 0.0001 10 10 146
matrix.47 ~/~ matrix.47 0.43 0.0078 < 0.0001 11 11 147
matrix.55 ~/~ matrix.55 0.45 0.0082 < 0.0001 12 12 148
rotate.3 ~/~ rotate.3 0.32 0.0057 < 0.0001 13 13 149
rotate.4 ~/~ rotate.4 0.32 0.0058 < 0.0001 14 14 150
rotate.6 ~/~ rotate.6 0.38 0.0068 < 0.0001 15 15 151
rotate.8 ~/~ rotate.8 0.33 0.0059 < 0.0001 16 16 152We can also see some information on the model fit, which can be used for making comparisons if we make any changes to the model (e.g., pruning).
Measure Value
logl -13298.10
unrestricted.logl -13298.10
baseline.logl -15949.45
nvar 16
nobs 152
npar 152
df ~ 0
objective -11.94
chisq ~ 0
pvalue 1
baseline.chisq 5302.71
baseline.df 120
baseline.pvalue ~ 0
nfi 1
pnfi ~ 0
tli
nnfi 1
rfi
ifi 1
rni 1
cfi 1
rmsea
rmsea.ci.lower ~ 0
rmsea.ci.upper ~ 0
rmsea.pvalue ~ 0
aic.ll 26900.19
aic.ll2 26934.10
aic.x ~ 0
aic.x2 304
bic 27710.32
bic2 27227.45
ebic.25 28131.75
ebic.5 28553.18
ebic.75 28890.33
ebic1 29396.05Another important feature of the ggm() function is that statistically insignificant edges can be removed to have a more compact model. In the following codes, we will eliminate edges that are not statistically significant at the alpha level of \(\alpha = .05\), after performing a Benjamini and Hochberg (known as “BH” or “fdr”) correction (Benjamini & Hochberg, 1995). We will then compare this model with the previous model.
# We can prune this model to remove insignificant edges
network_sapa_5 <- psychonetrics::ggm(data = sapa,
omega = "full", # or "empty" to set all elements to zero
delta = "full", # or "empty" to set all elements to zero
estimator = "FIML", # or "ML", "ULS", or "DWLS"
verbose = FALSE) %>%
psychonetrics::runmodel() %>%
psychonetrics::prune(adjust = "fdr", alpha = 0.05)
# Compare the models
comparison <- psychonetrics::compare(
`1. Original model` = network_sapa_4,
`2. Compact model after pruning` = network_sapa_5)
print(comparison) model DF AIC BIC RMSEA Chisq
1. Original model 0 26900.19 27710.32 ~ 0
2. Compact model after pruning 73 26985.28 27406.33 0.038 231.09
Chisq_diff DF_diff p_value
231.09 73 < 0.0001
Note: Chi-square difference test assumes models are nested.The results show that the compact model with pruning fits the data better based on BIC although AIC shows the opposite result. In the final step, we will visualize the last network model resulting from ggm() using the famous qgraph package (Epskamp et al., 2012):
library("qgraph")
# Obtain the network plot
net5 <- getmatrix(network_sapa_5, "omega")
qgraph::qgraph(net5,
layout = "spring",
theme = "colorblind",
labels = colnames(sapa))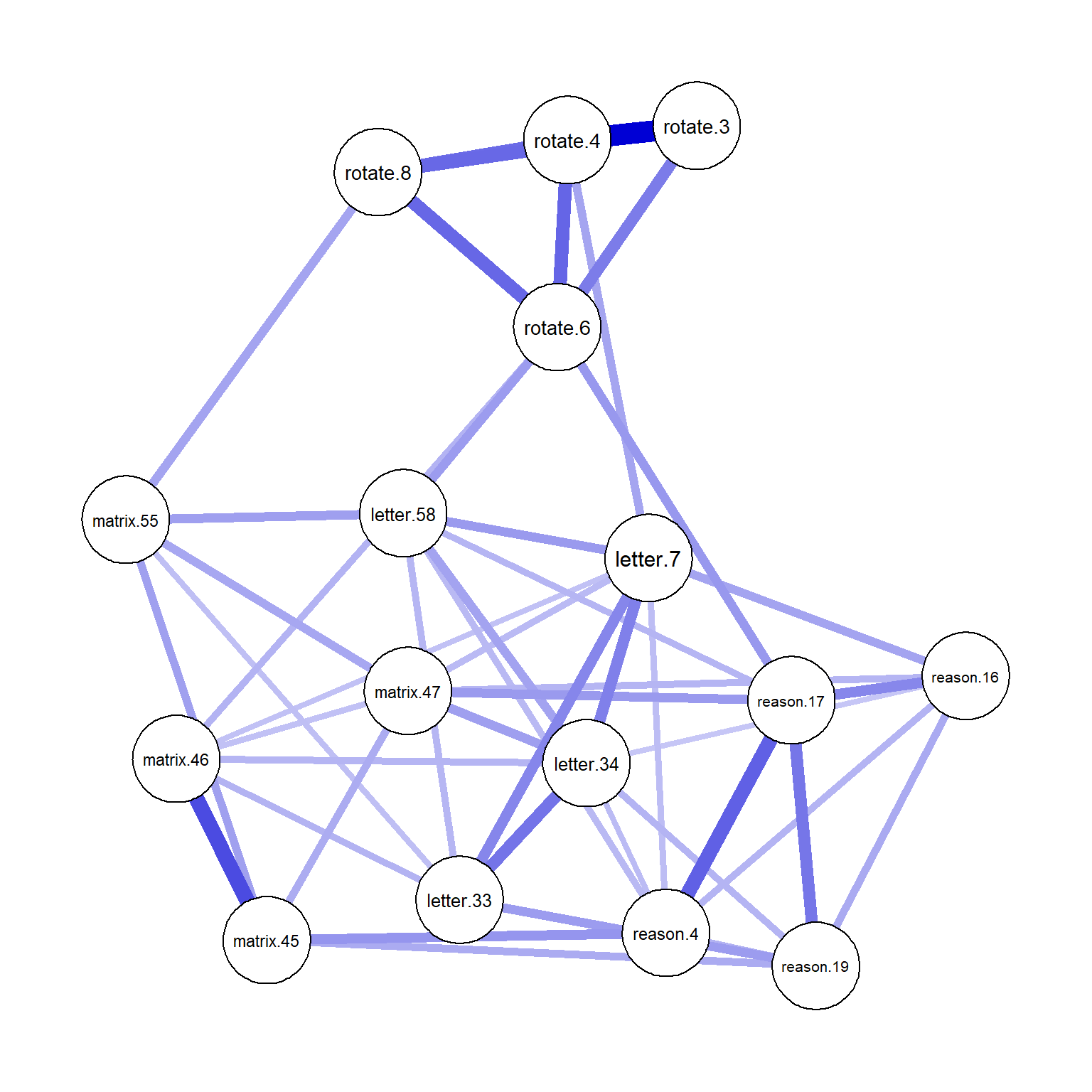
Figure 7: Network plot using qgraph
Conclusion
This was a very brief introduction to PNA using R. There are still many things to discuss in PNA. Thus, I plan to write several blog posts on PNA over the next few months. For example, I want to discuss centrality indices and demonstrate how to generate centrality indices for network models using R. I will also write a post on the Ising model for binary data to demonstrate the similarities and differences between an Ising network model and an item response theory (IRT) model.